Call:
lm(formula = Petal.Width ~ Sepal.Length, data = iris)
Residuals:
Min 1Q Median 3Q Max
-0.96671 -0.35936 -0.01787 0.28388 1.23329
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) -3.20022 0.25689 -12.46 <2e-16 ***
Sepal.Length 0.75292 0.04353 17.30 <2e-16 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 0.44 on 148 degrees of freedom
Multiple R-squared: 0.669, Adjusted R-squared: 0.6668
F-statistic: 299.2 on 1 and 148 DF, p-value: < 2.2e-16Biostat 823 - Literate Programming
Duke University, Department of Biostatistics & Bioinformatics
2024-08-29
Literate Programming
First introduced by Donald Knuth (“The Art of Computer Programming”) in 1984
Let us change our traditional attitude to the construction of programs: Instead of imagining that our main task is to instruct a computer what to do, let us concentrate rather on explaining to human beings what we want a computer to do.
D. E. Knuth, Literate Programming, The Computer Journal, Volume 27, Issue 2, 1984, Pages 97–111, https://doi.org/10.1093/comjnl/27.2.97
TANGLE and WEAVE
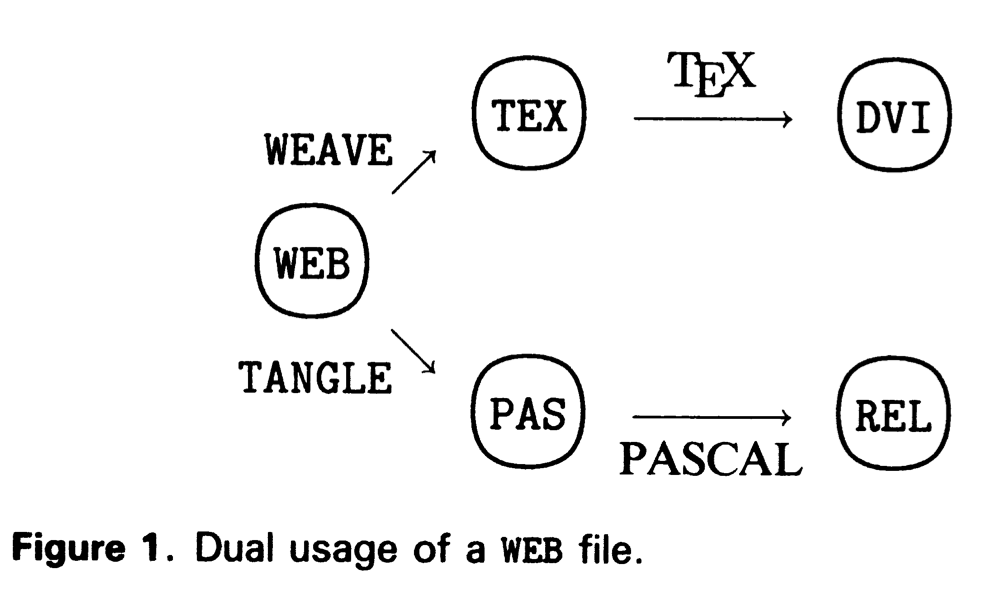
Figure 1 from Knuth (1984)
Lit. Prog. and Reproducible Research
Literate Programming: Enhances traditional software development by embedding code in explanatory essays and encourages treating the act of development as one of communication with future maintainers
Reproducible Research: Embeds executable code in research reports and publications, with the aim of allowing readers to re-run the analyses described.
Code not paper is the scholarship
An article about computational science in a scientific publication is not the scholarship itself, it is merely advertising of the scholarship. The actual scholarship is the complete software development environment and complete set of instructions which generated the figures.
Research Compendium
- Encapsulates the actual work, not just an abridged version;
- Allows different levels of detail in different renderings;
- Easy to re-run by anyone;
- Provides explicit computational details, enabling others to adapt and extend the reported computational methods;
- Enables programmatic construction and clear provenance of plots and tables;
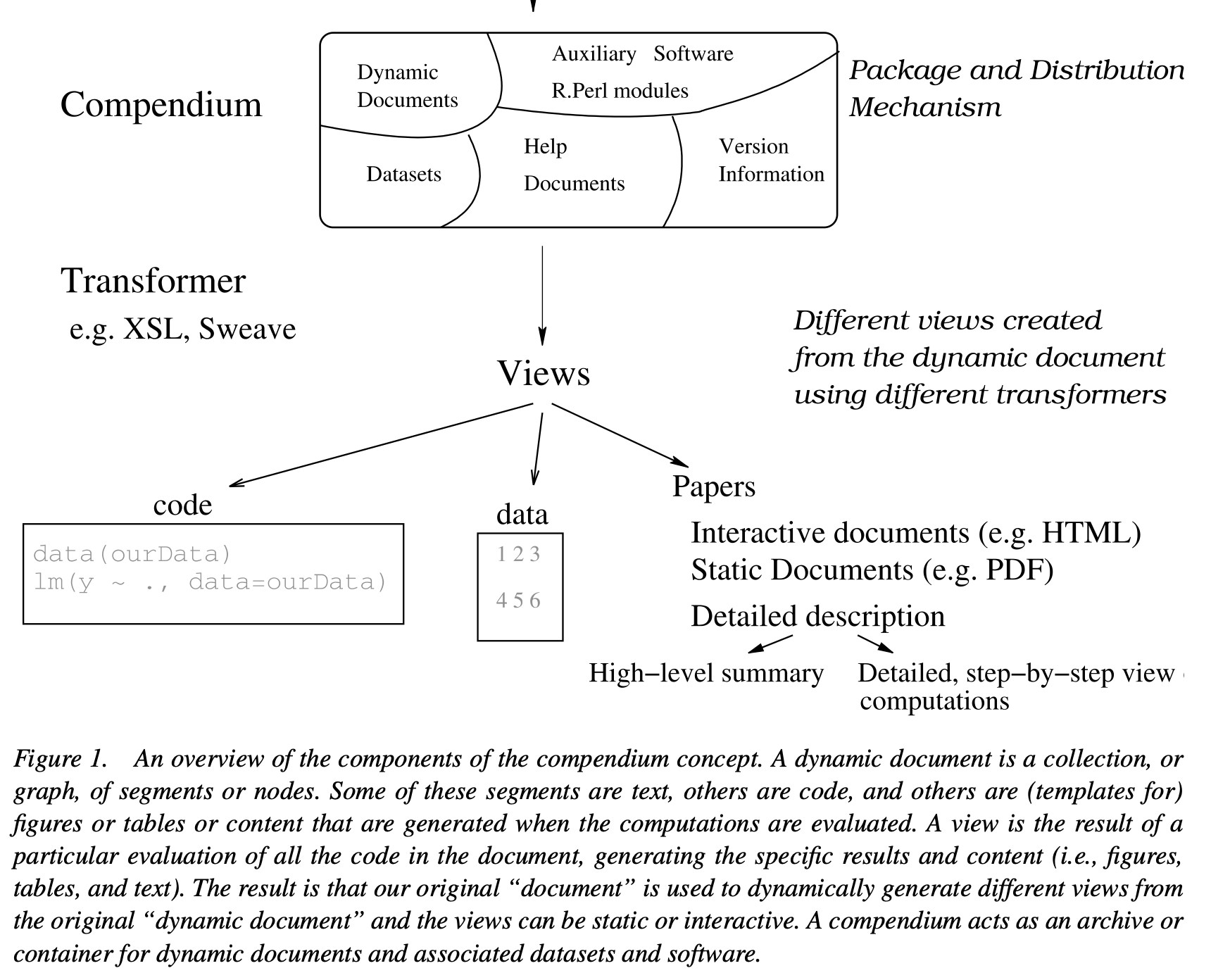
Part of Figure 1, Gentleman and Temple Lang (2007)
Reproducible research lens
Embeds executable code in research reports and publications, with the aim of allowing readers to re-run the analyses described. (Schulte et al (2012))
Important concepts:
- Ties together narrative (the “why”), code that implements it, and the results from running the code.
- Can be executed (“executable paper”), in its original or modified form.
- Provenance of tables and charts is clear and verifiable.
Sweave (Leisch 2002)

Part of Figure 1, F. Leisch (2002). “Sweave, Part I: Mixing R and LaTeX: A short introduction to the Sweave file format and corresponding R functions”. R News. 2 (3): 28–31.
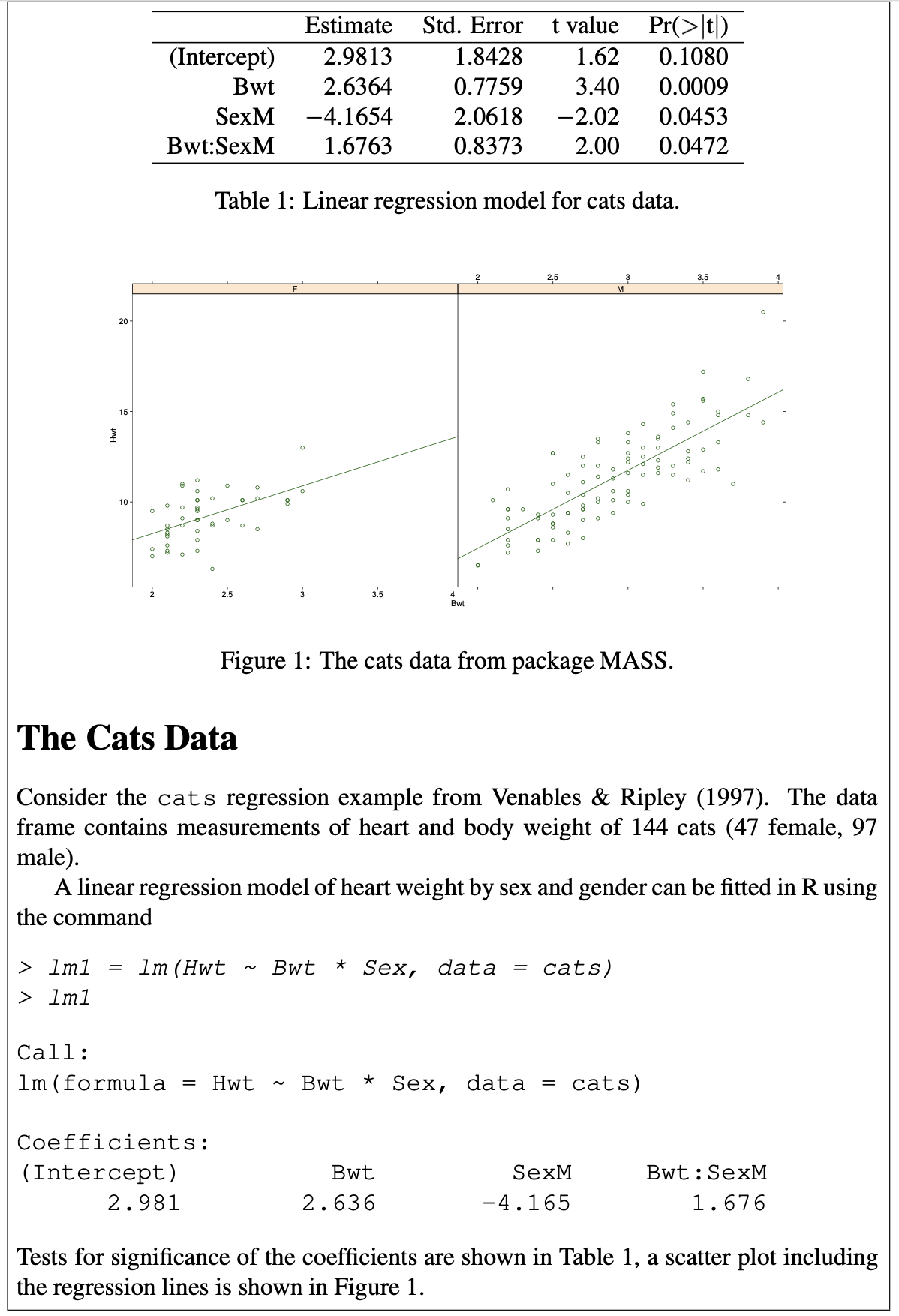
Figure 2, F. Leisch (2002)
Renewed interest for reproducible computing
- The practice of copying values for tables and plots for figures into a document breaks the provenance chain.
Provenance: Information about entities, activities, and people involved in producing data or other results, which are necessary to assess their quality, reliability or trustworthiness.1
“Executable Paper” concept:
- eLife “Welcome to a new ERA of reproducible publishing”
- For example: L Michelle Lewis et al, Reproducibility Project: Cancer Biology (2018) Replication Study: Transcriptional amplification in tumor cells with elevated c-Myc. eLife 7:e30274. Executable version
- Boettiger C et al (2016). “RNeXML: A Package for Reading and Writing Richly Annotated Phylogenetic, Character, and Trait Data in R.” Methods in Ecology and Evolution, 7, pp. 352-357. Rmarkdown version and rendered PDF
Jupyter and iPython Notebooks
- iPython (interactive Python) created by Fernando Pérez in 2001, gained Notebook interface in 2011
- Project Jupyter started in 2015 as a spin-off
- Supports a multitude of kernels
- JupyterLab first released in 2018
- JupyterHub first released in 2019
- Intro to Jupyter1
- Itself generated from a source Jupyter Notebook
Knitr and Rmarkdown
- Knitr created by Yihui Xie, first released 2012

- Designed as a general-purpose literate programming engine
- Design allows different input languages and different output formats
- Rmarkdown
- First introduced in knitr in early 2012, withh the idea to embed code chunks in Markdown documents.
- rmarkdown R package created in 2014
- Rich universe of examples at RPubs
Markdown and code
In Rmarkdown, code can be inline:
Consider Edgar Anderson's Iris data of sepal and petal lengths measurements
of `r nrow(iris)` flowers, (`r sum(iris$Species=="setosa")` _I. setosa_,
`r sum(iris$Species=="versicolor")` _I. versicolor_, and
`r sum(iris$Species=="virginica")` _I. virginica_).Consider Edgar Anderson’s Iris data of sepal and petal lengths measurement of 150 flowers, (50 I. setosa, 50 I. versicolor, and 50 I. virginica).
Code chunks
A linear regression model of petal width by sepal length can be fitted in the following way:
Rendered:
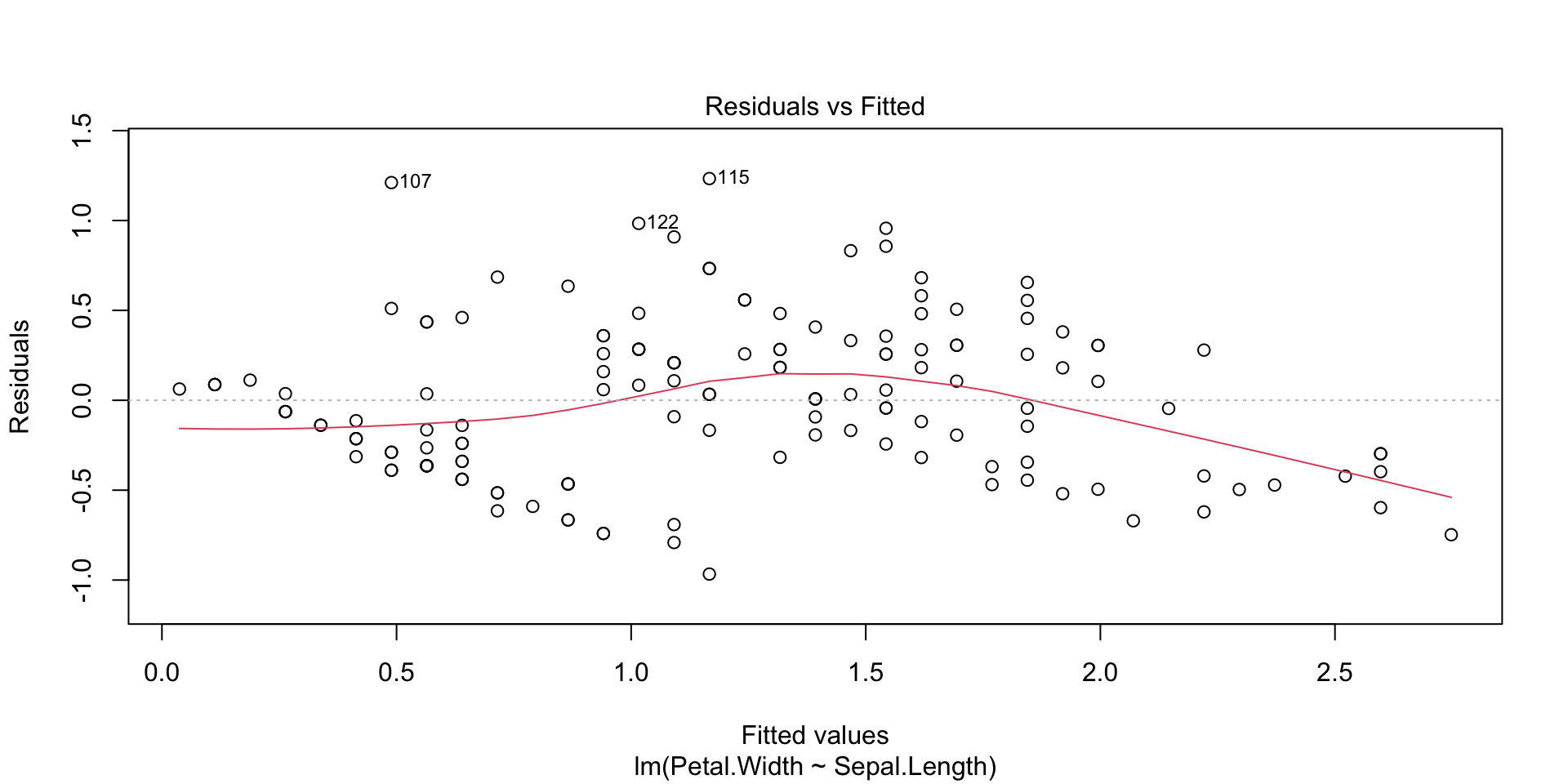
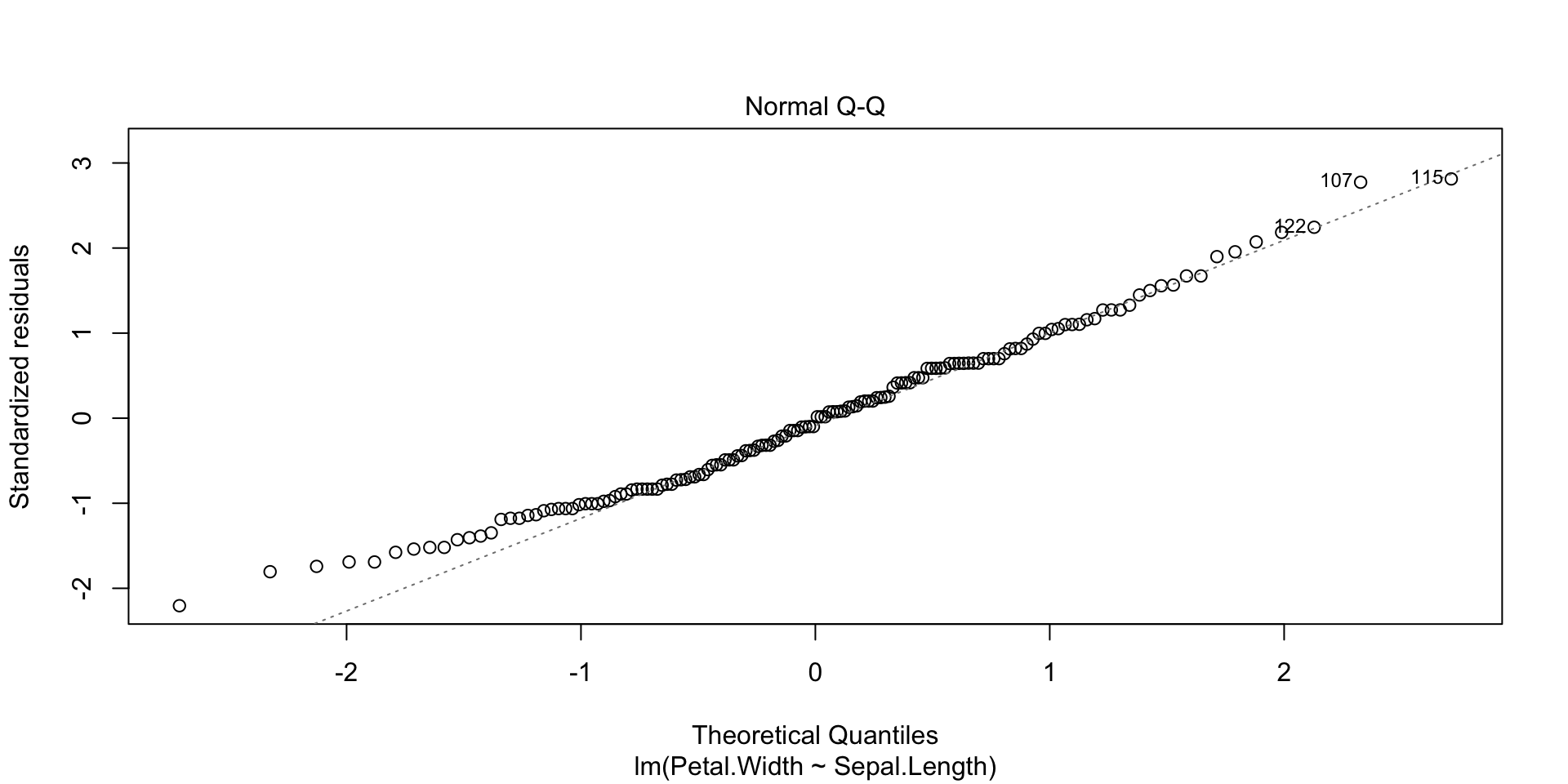
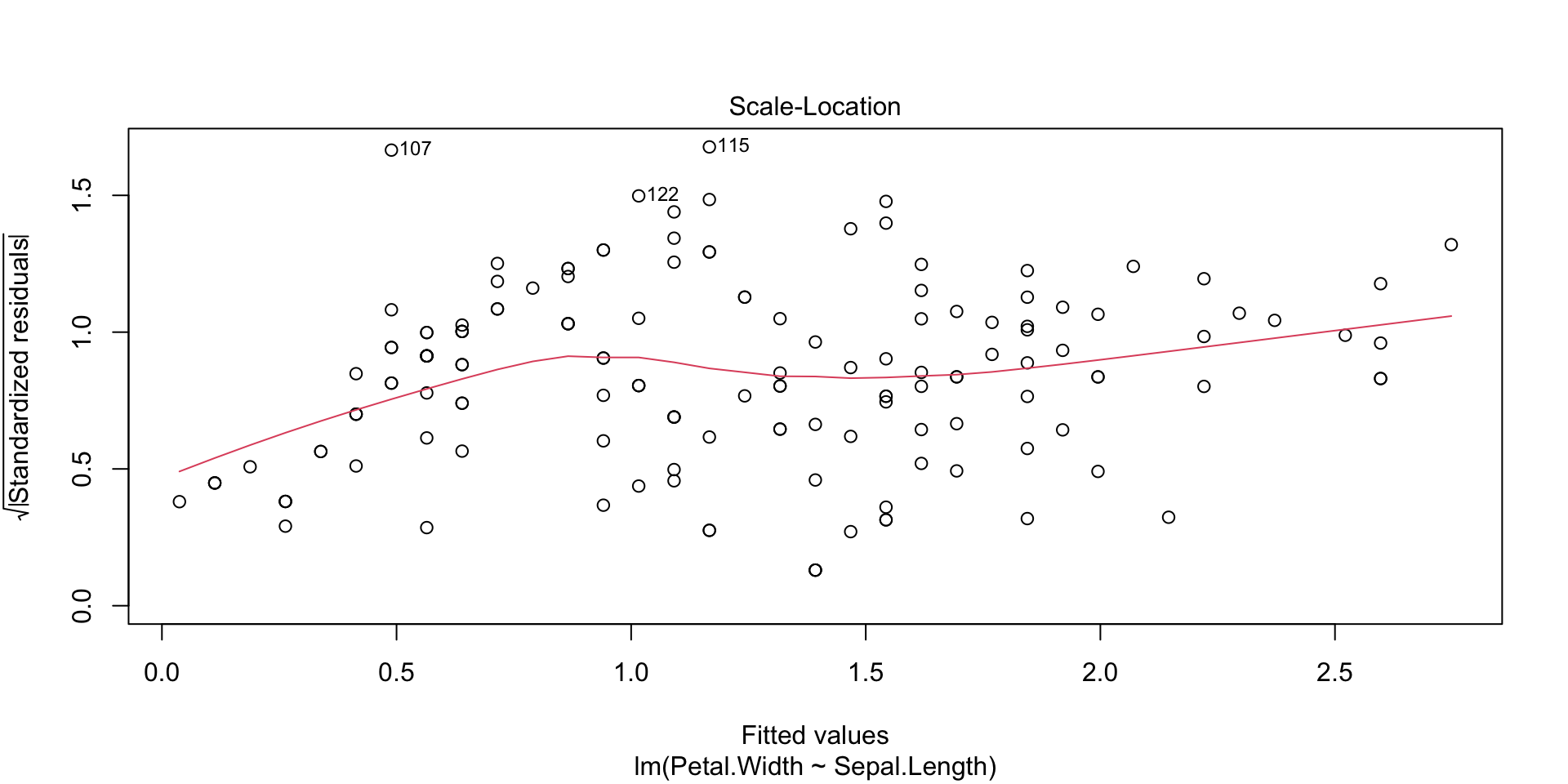
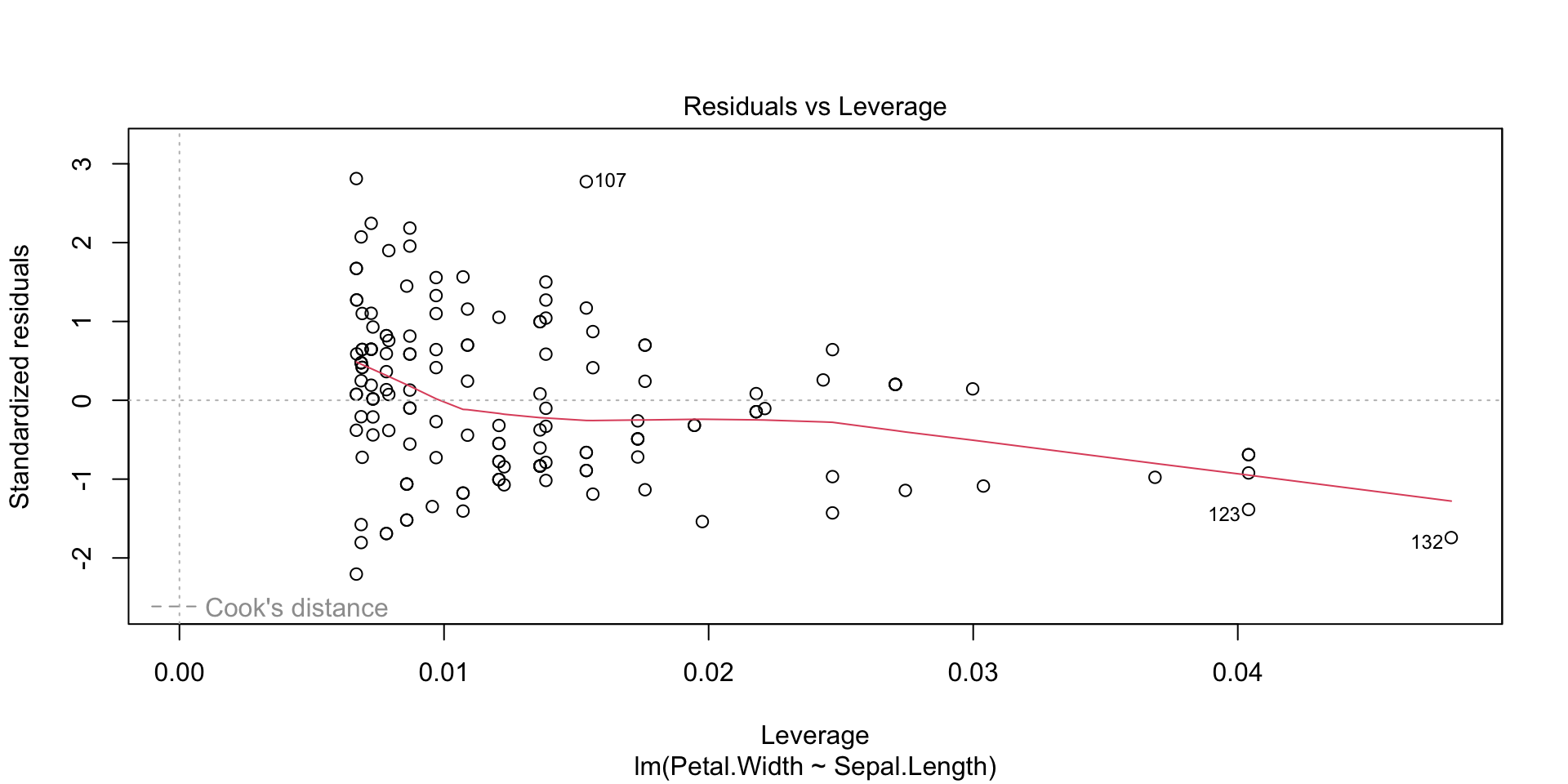
Plots also get inlined
Plots of the regression residuals etc can be obtained in the following way:
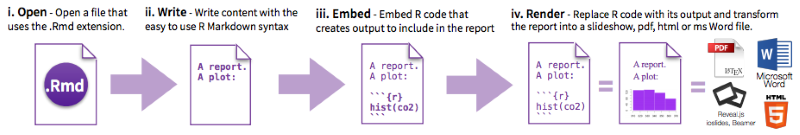
Ecosystem
One report document, rendered to many different output formats
Further reading
- List of Literate Programming environments on Wikipedia
- Xie Y, Allaire J, Grolemund G (2018). R Markdown: The Definitive Guide. Chapman and Hall/CRC, Boca Raton, Florida. ISBN 9781138359338
- Resul Umit (2022) Writing Reproducible Research Papers with R Markdown
- Chapters “Jupyter Notebook for Open Science”, “R for Reproducible Scientific Analysis (Jupyter)”, and “R for Reproducible Scientific Analysis (RMarkdown / knitr)” in Reproducible analysis and Research Transparency Workshop (2017)
- J.M. Perkel (2018). Why Jupyter is data scientists’ computational notebook of choice. Nature 563, 145-146. doi:https://doi.org/10.1038/d41586-018-07196-1
- Rule, Adam, Amanda Birmingham, Cristal Zuniga, Ilkay Altintas, Shih-Cheng Huang, Rob Knight, Niema Moshiri, et al. 2019. “Ten Simple Rules for Writing and Sharing Computational Analyses in Jupyter Notebooks.” PLOS Computational Biology 15 (7): e1007007. https://doi.org/10.1371/journal.pcbi.1007007