Audience: Many Different Backgrounds¶
- New to programming, never seen any code before
- Have done some programming, but not confident with the subject
- Extensive programming, confident with the subject
- Former software engineer, professional
Practice Practice Practice!¶
- Just like learning how to ride a bike, you start by doing
- No wrong choice on what programming language to start with or learn
- What is important are the fundamental concepts for writing code, not the specifics of the language
- Lots of concepts are important in any programming language you want to use
Why Python?¶
- It is a general purpose programming language popular among many disciplines
- Ex/ scientific computing, software engineering, finance
- Large community among scientists
- It’s free, well-documented, and runs everywhere
- Relatively quick to start using, but there is a lot to learn!
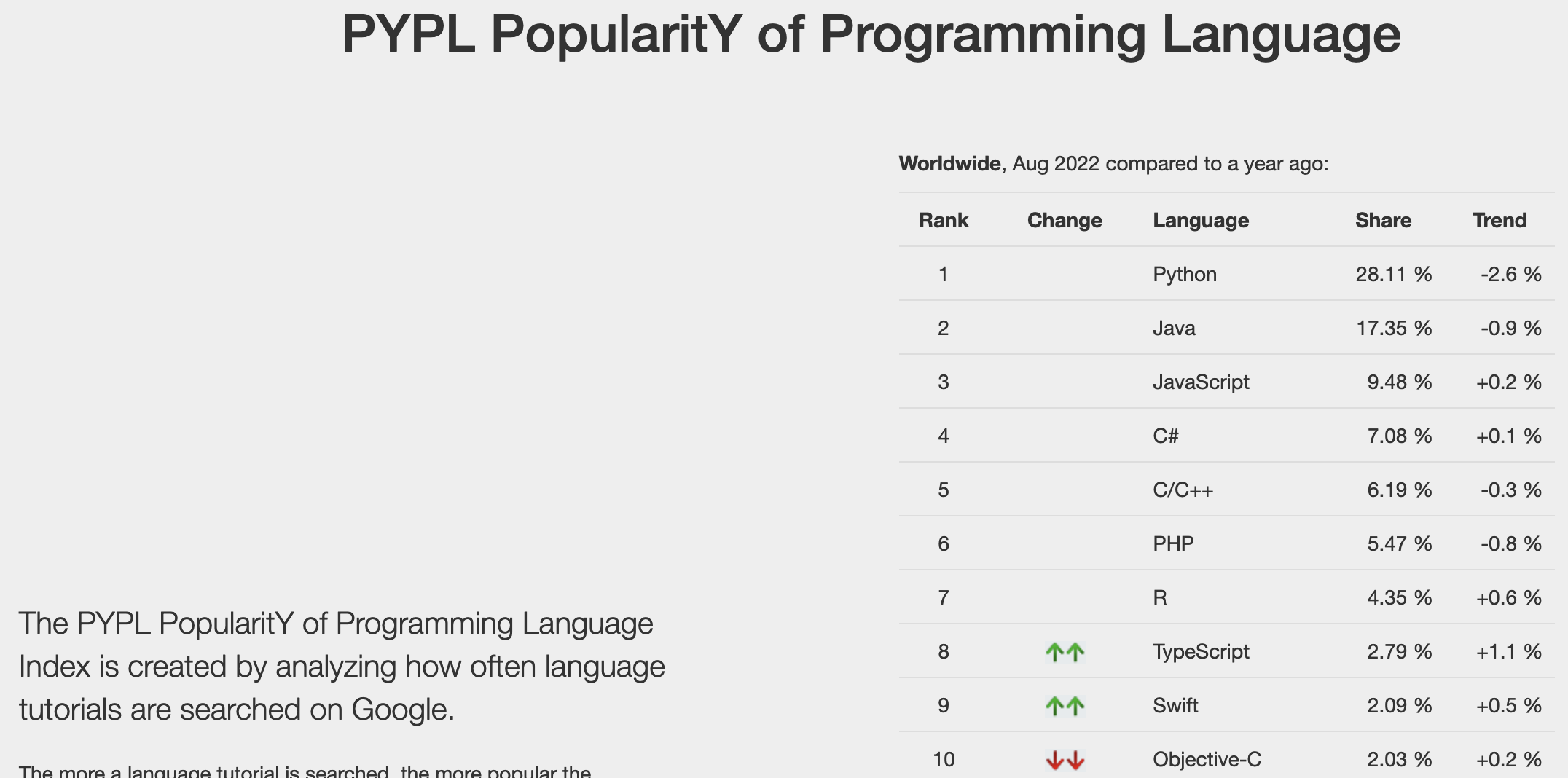
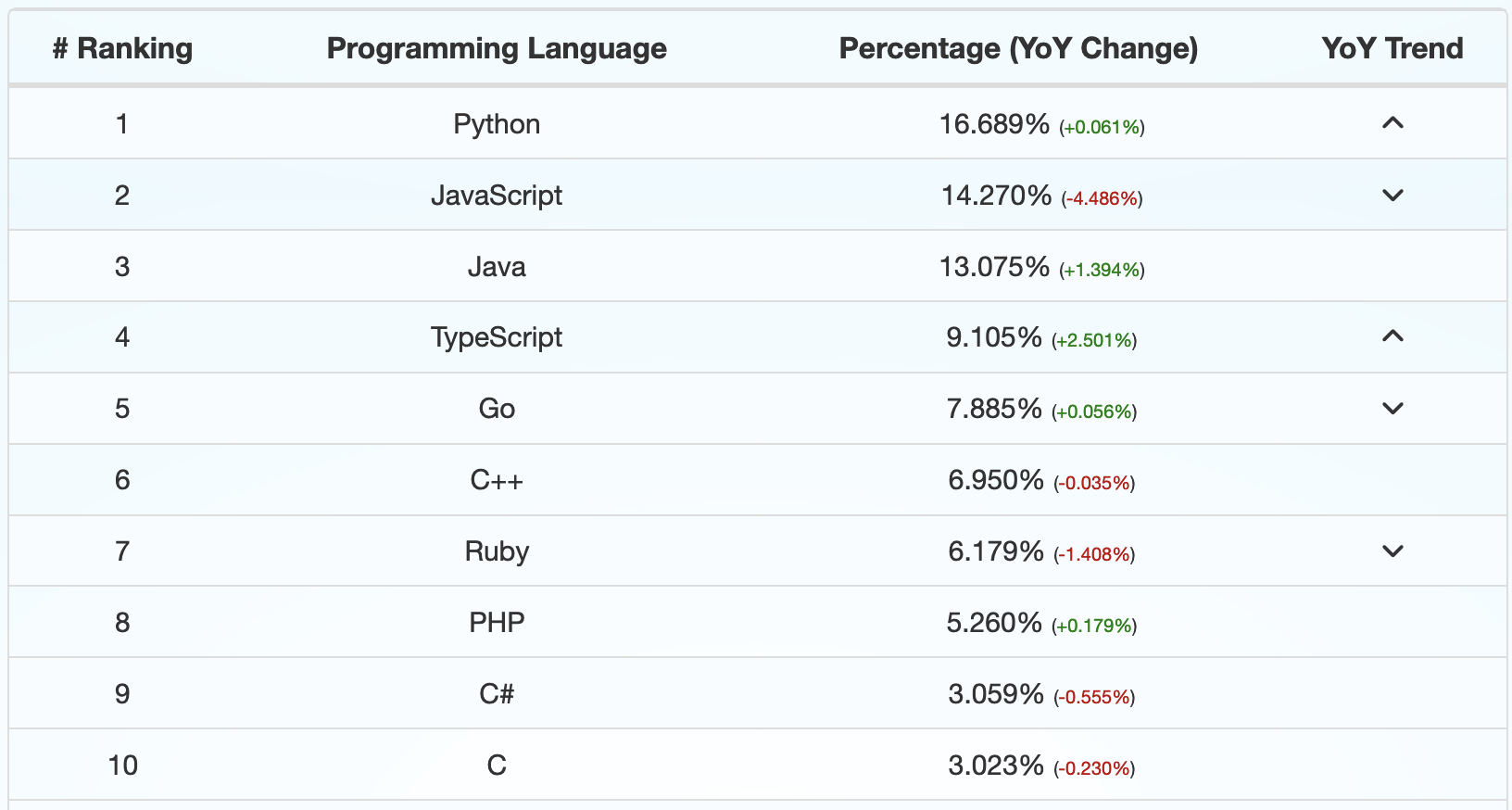
Percent of pull requests on GitHub by language in Q1 of 2022.¶
Data from: https://madnight.github.io/githut/#/pull_requests/2022/1
Goals: Write and Run Programs¶
- Cover common data types and functions
- Making decisions in a program with if statements
- Writing for loops to apply code to a group of data
- Writing our own functions
- Work with files and libraries
- Know where to look for more help
Expectations¶
- This is alot of material
- Especially for someone new to programming
- I will do my best to go through the material at an appropriate pace
- Feel free to let me know if I am:
- Going too fast
- Need to explain something in more detail
- Provide more examples
- etc.
Expectations¶
- If you are having technical issues, use a sticky note
- This will be taught with live coding, I will probably have technical issues
- Feel free to ask me questions
- You can also use a sticky note to signal a helper or another instructor to help you with a question
- Or ask a neighbor
Begin Jupyter Notebook¶
Recap and Exercise: Data Types and Variables¶
Recap: Loops, Dictionaries, Lists, and Conditionals¶
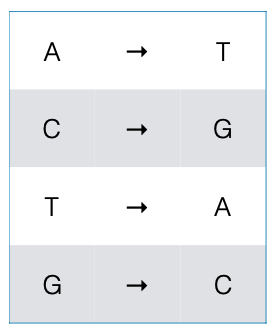
Complementing¶
- We can loop over all the bases in a sequence
- Each base has a complement that we should substitute
- We can use a Dictionary to store this mappping.
Dictionaries and Lists¶
Create dictionaries with {}, lists with []
nucs = {'A': 5, 'C': 4, 'T': 8}
counts = [5,4,8]Both accessed with [] - dictionaries by key, lists by index
nucs['A'] # 5
counts[0] # 5
nucs['A'] = 3 # now 3
counts[0] = 3 # now 3GC-content percentage¶
- Calculated as (G + C) / (A + T + G + C)
- Create a GC count variable and an ATGC count variable
- Loop over each base in the sequence
- If G, add 1 to GC count
- If C add 1 to GC count
- For everything, add 1 to ATGC count
- Loop over each base in the sequence
Conditionals¶
# Test c1 for True or False
if c1:
print("c1 was True")
# c1 was False, check c2
elif c2:
print("c1 False but c2 True")
# All checks False
else:
print("Both False")Exercise: Using Functions¶
bases = 'adenine cytosine guanine thymine' Write some code that:
- Makes a list of these bases from the string
- Uppercases the names (e.g. ['ADENINE', ...])
- Reverses the order (e.g. ['THYMINE',...])
Hint: Use help(str) and help(list) to see what functions are available for strings and lists
Bonus: Write a for loop to print the first letter of each (e.g. A, C, ...)
Exercise: Update the Reverse Function¶
Strings can be reversed with this special slicing notation: [::-1]
s = 'abc'
r = s[::-1]
print(r)cba
Update reverse() function to use [::-1] instead of a loop.
Do we need to do anything to complement()?
What about reverse_complement()?
Recap and Exercise: Making Functions and Reading Files¶
Functions¶
Calling functions: length = len('abc')
- Defining functions:
def double(x): return x * 2 - Composing functions:
def reverse_complement(seq): return reverse(complement(seq))
Avoid using global variables in functions
Reading files¶
- Open a file with the
open()function:f = open('ae.fa') - Loop over lines, and
strip()each onefor line in f: print(line.strip()) - Close with
f.close()
Exercise: Reading a Fasta File¶
- Write a function,
read_fasta(filename)that:- Takes 1 argument:
filename - Reads the file line-by-line
- Strips/combines the lines into one long line
- Skips the line if it contains a '>' character
- Takes 1 argument:
Scripts¶
- Put code in a file, give it the .py extension
- Read command line-arguments from sys.argv:
import sys
print(sys.argv[0])
print(sys.argv[1])$ python script.py hello
script.py
hello- Check the length of sys.argv to be helpful!
Summary¶
- We introduced several data types inculding:
- Integers, Floats, Strings, Dictionaries, Lists
- How to assign values to variables and use them
- Making choices using conditional statements
- Writing for loops to perform a task over a group of data
- Making our own functions
- Work with files and libraries
- Help function:
help()